This is the Apache Iceberg deep dive associated with the Table format comparisons - Change queries and CDC blog post, that looks at how each table format supports change queries tables, including full CDC. This is not a how-to guide but an examination of Iceberg capabilities through the lens of its internal design.
Preamble
In the parent blog post, I described four canonical change query types:
latest-state
append-only: Return all inserted rows covering the query time period.
upsert: Return all inserted and updated rows (without the before image) covering the query time period. This may or may not return rows that were deleted in the time period; this will depend on the table format.
CDC
min-delta: Minimized CDC results covering the query time period.
full-delta: Complete CDC results, including redundant changes, covering the query time period.
I took the append-only and min-delta names from this incremental processing Snowflake paper that describes how Snowflake change queries work. Upsert and full-delta are additional types that are not supported by Snowflake but have some support among the table formats. We’ll be framing the Iceberg capabilities in terms of those four change query types.
In the parent post I also covered the three main approaches to CDC support:
Materialize CDC files on write
Infer-CDC-on-read
Row-lineage (aka row tracking)
The most important thing to know about Iceberg and change queries is that it does not have native CDC support today, so compute engines must implement their own infer-CDC-on-read implementation. I’ll cover how compute engines can theoretically implement CDC queries and how Apache Spark does it today, including its limitations. We’ll also look at row-lineage which may hit Iceberg v3, which is the basis for how Snowflake implements append-only and min-delta.
Some relevant internals background
The internals of a table format determine how change queries are implemented and can also determine the limitations in these early, still-maturing technologies. They may have been around for a few years now, but we’re still in the early phase, where a lot of the features are still being added at a fast pace.
The following sections describe some aspects of Iceberg internals that are important for understanding the current state of change queries in Iceberg.
Iceberg snapshot types
Iceberg has different types of operations, and the operation type of each commit is logged as a field in its snapshot. As an Iceberg user, you shouldn’t normally need to know this stuff… but abstractions leak.
The snapshot types (known as operation types in the code) that are relevant here are:
Append: A transaction only adds data files. In the context of SQL, these correspond to inserted rows from INSERT INTO commands, bulk inserts or streaming appends from Flink.
Overwrite: Used for row-level inserts, updates and deletes. A commit may include both the addition and deletion of data files, and delete files (if merge-on-read is used). Corresponds to MERGE, UPDATE, and DELETE commands.
Delete: Only deletes data files. In the context of SQL, these correspond to DELETE commands that align with partitions to delete whole data files at a time rather than a subset of rows (which would be an Overwrite commit).
Replace: A compaction job replaces a set of data and delete files with a new set of data files. Logically the data remains unchanged and so none of the compute engines pay attention to these commits for incremental latest-state or CDC reads.
These snapshot types are relevant to the current limitations of Iceberg support for change queries.
Incremental table scans
The Iceberg core module offers compute engines three ways of performing incremental table scans:
Data file iterators:
Table.IncrementalAppendScan: A compute engine uses this method to obtain an incremental scan that acts as a data file iterator over Append snapshots. This type of scan can be used to read inserted rows.
Table.IncrementalChangelogScan: A compute engine obtains an incremental scan that acts as a data file iterator over Append, Overwrite, and Delete snapshots. It does not currently support delete files so this method is best for copy-on-write (COW) tables.
A compute engine can walk the log of snapshots directly using the SnapshotUtil class, discovering the added and removed data/delete files in each snapshot as it goes. This approach is the most flexible as it gives the compute engine access to the snapshots, rather than just a data file iterator.
From there, it’s up to the compute engine to use these iterators to perform incremental change queries.
Infer-CDC-on-read with copy-on-write vs merge-on-read
Given Iceberg does not offer CDC file materialization or row-lineage yet; how can a compute engine know which rows have been inserted, updated and deleted?
It depends on whether a table uses copy-on-write (COW) or merge-on-read (MOR), or both. We’ll ignore mixed mode tables for clarity.
Copy-on-write (COW)
In a COW table, an update or delete of an existing row will cause a data file to be rewritten with the change applied. The new data file may have a number of:
New rows that didn’t exist in the original data file.
Carried-over (unchanged ) rows from the original data file.
Updated rows that exist in their original form in the original data file and in their new form in the new data file.
Absent rows that exist in the original data file but don’t exist in the new data file.
Fig 1. The deleted and added data files of snapshot S are compared to yield a CDC result.
An UPDATE, DELETE or MERGE operation could create multiple pairs of deleted and added data files. When a snapshot includes multiple added and deleted data files, the compute engine cannot know which are the add/delete file pairs, and therefore must do a row-level comparison based on all of the added/deleted data files of the snapshot.
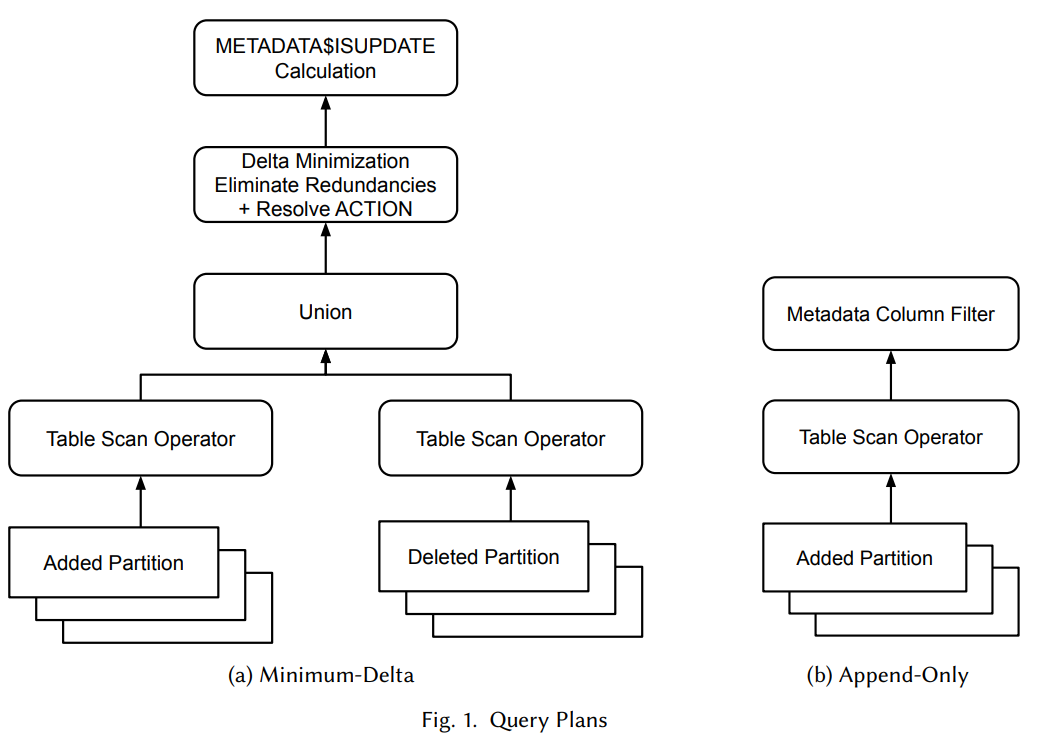
Fig 2. Taken from the Snowflake paper: What’s the Difference? Incremental Processing with Change Queries in Snowflake. Snowflake tables use copy-on-write (COW). Shows a Snowflake min-delta query plan that results in the rows returned from scanning multiple added data files (known as partitions in Snowflake) and multiple (logically) deleted data files being unioned into a diffing phase. A similar approach is required for Iceberg COW tables.
A MERGE command can insert, update, and delete rows in one operation. An example of a merge statement that produces the CDC result of fig 1 is seen in fig 3 below.
Fig 3. A merge statement synchronizes a source and target table.
This row-level diff between data files is the only way to do infer-CDC-on-read with Iceberg COW tables.
Merge-on-read (MOR)
In a MOR table, data files are not logically deleted and rewritten. Instead:
New and updated rows are written to new data files
The original locations of updated rows are invalidated with a delete file entry.
Deleted rows are invalidated with a delete file entry.
Fig 4. The added data file and delete file of snapshot S, as well as the referenced data file of the two delete file entries, are read to yield a CDC result.
To infer the changes that a snapshot has made with MOR is less IO and computationally expensive. Now the compute engine can:
Provisionally treat all rows in added data files as either inserts or updates. There are no carry-over rows to be ignored, and the data files are smaller requiring less IO.
If no delete file was added, then all rows of the data file are inserts.
If a delete file was added, then it can tell the compute engine which (if any) of the rows in the added data file are updates, and also where to find the rows of existing data files that are deletes. The referenced data files must be read to get the delete row state, but at least it doesn’t require costly row-level diffing.
It’s still more costly than reading materialized CDC files, but it's better than COW.
That’s enough internals background; let’s dive into what you can do with Iceberg today.
Append-only and Upsert Change Query support
As already described there are three methods for a compute engine to incrementally read an Iceberg table. The following describes how Flink, Spark and Spark Structured Streaming use these methods:
Apache Flink supports consuming a mutable table as a streaming source using the IncrementalAppendScan approach, which corresponds to append-only change queries. In the append-only tables blog post, I covered how Apache Flink incrementally reads append-only tables, and it is this same mechanism but applied to mutable tables. While it corresponds to the append-only query type, it does not entirely conform to our definition, which defines it as returning all inserts in the time period. The IncrementalAppendScan only iterates over the data files of Append snapshots, and therefore, row inserts from MERGE commands are skipped (as merges create Overwrite snapshots). Therefore, it is append-only but is not guaranteed to read all inserts.
Apache Spark allows you to perform incremental change queries by supplying a starting timestamp or snapshot ID for the query. With a starting point, Spark uses the same IncrementalAppendScan approach as Flink. This results in exactly the same, not quite conformant append-only change query.
Spark Structured Streaming uses the snapshot iteration approach. When you kick off a readStream operation, SSS will perform a batch scan to return the current state of the table based on the starting snapshot (supplied by the user or defaults to the current snapshot). Then it runs periodic incremental scans that iterate through the subsequent snapshots to return the changes that occur after that starting point. The incremental portion does not support Overwrite or Delete commits and will error if it encounters one. If you want to avoid the streaming read from erroring, you must use the streaming-skip-overwrite-snapshots and streaming-skip-delete-snapshots configs to skip those commits entirely. The result is that SSS like Flink and Spark can only read Append commits.
I haven’t looked at other compute engines, but of the three sampled, they all:
Offer the same limited append-only change query support.
No support for upsert change queries.
Iceberg isn’t alone with this behavior, so don’t think it’s only Apache Iceberg with such a limitation. Delta Lake has similar but different limitations, as we’ll see in the next analysis when it comes out. I haven’t tested Snowflake, but from the linked paper, it seems that Snowflake will return rows inserted by MERGE statements. Snowflake can do this at low cost due to row lineage, which we’ll look at briefly later on.
CDC change query support
Iceberg does not yet have native support for CDC but that hasn’t stopped Spark from implementing its own infer-CDC-on-read implementation. I will cover that support in this section.
Iceberg supports views. Generically, a view is a logical table that doesn’t contain any data but acts like a stored query that you can query by name. Views are common in regular OLTP database tables and Iceberg supports them as well. Spark has a procedure called create_changelog_view that adds a changelog view to an Iceberg catalog.
When you query an Iceberg changelog view, Spark executes a CDC change query by using the IncrementalChangelogScan of the Iceberg core module that returns added and logically deleted files of Append, Overwrite, and Delete snapshots. The main limitation of this scan is that it doesn’t return delete files making it unsuitable for MOR tables. If you try and use a changelog view in Spark on a MOR table, it will error.
Spark provides a number of configs:
For incremental reads use either snapshot or timestamp based options:
start-snapshot-id
end-snapshot-id
start-timestamp
end-timestamp
net_changes: When true, it returns a minimized CDC result. When false, it returns a full CDC result, including redundant changes.
compute_updates: Whether to include pre and post images of updates. When false, an update will be registered as a delete and insert pair.
identifier_columns: Tells Spark how to identify each row, like a primary key which is necessary to compute the pre and post images of updates.
Let’s go through some examples using Pyspark with an Iceberg table configured to use copy-on-write. First, we’ll insert three rows.
spark.sql("""
INSERT INTO local.fruit
VALUES ('jack', 'apple'), ('sarah', 'orange'), ('john', 'pineapple')
""")
Then we’ll perform an update and then a delete.
spark.sql("""
UPDATE local.fruit
SET fruit = 'banana'
WHERE name = 'jack'
""")
spark.sql("""
DELETE FROM local.fruit
WHERE name = 'john'
""")
Next we’ll create two changelog views, one for a minimized CDC feed and another for a complete CDC feed.
spark.sql("""
CALL local.system.create_changelog_view(
table => 'local.fruit',
changelog_view => 'fruit_min_delta',
net_changes => true
)
""")
spark.sql("""
CALL local.system.create_changelog_view(
table => 'local.fruit',
changelog_view => 'fruit_full_delta',
identifier_columns => array('name'),
compute_updates => true
)
""")
Now we can query each to see the results. First the complete CDC change query.
spark.table("fruit_full_delta").sort(array('_change_ordinal'))
.show()
+-----+---------+-------------+---------------+-------------------+
| name| fruit| _change_type|_change_ordinal|_commit_snapshot_id|
+-----+---------+-------------+---------------+-------------------+
| jack| apple| INSERT| 0| 260123709935954149|
|sarah| orange| INSERT| 0| 260123709935954149|
| john|pineapple| INSERT| 0| 260123709935954149|
| jack| apple|UPDATE_BEFORE| 1|1410375155145569705|
| jack| banana| UPDATE_AFTER| 1|1410375155145569705|
| john|pineapple| DELETE| 2|6803657404732109534|
+-----+---------+-------------+---------------+-------------------+
Next, the minimized CDC change query.
spark.table("fruit_min_delta").sort(array('_change_ordinal'))
.show()
+-----+------+------------+---------------+-------------------+
| name| fruit|_change_type|_change_ordinal|_commit_snapshot_id|
+-----+------+------------+---------------+-------------------+
|sarah|orange| INSERT| 0| 260123709935954149|
| jack|banana| INSERT| 1|1410375155145569705|
+-----+------+------------+---------------+-------------------+
Notice that the John row doesn’t appear at all in the minimized CDC results as it was inserted and then deleted - this was a redundant change and therefore excluded.
Both of these views are computed over all changes that exist in the log of snapshots in the current metadata file. Depending on how many snapshots are maintained in the log this is going to be expensive. It’s better to query this data incrementally.
If we only want an incremental change query that includes the changes since the last snapshot that we read up to, then we need to create a view that is bounded by a start and end snapshot id, or pair of timestamps.
You can specify the upper and lower bound snapshot ids by adding an options map to the view creation command:
options => map('start-snapshot-id','260123709935954149','end-snapshot-id', '6803657404732109534')
The following shows the results from:
Create a changelog view that includes the first snapshot, and query it.
Create another changelog view that includes the remaining 2 snapshots, and query it.
Create and query a changelog view covering the first snapshot +-----+---------+------------+---------------+-------------------+ | name| fruit|_change_type|_change_ordinal|_commit_snapshot_id| +-----+---------+------------+---------------+-------------------+ | jack| apple| INSERT| 0| 260123709935954149| | john|pineapple| INSERT| 0| 260123709935954149| |sarah| orange| INSERT| 0| 260123709935954149| +-----+---------+------------+---------------+-------------------+ Create and query a changelog view covering the last two snapshots +----+---------+-------------+---------------+-------------------+ |name| fruit| _change_type|_change_ordinal|_commit_snapshot_id| +----+---------+-------------+---------------+-------------------+ |jack| apple|UPDATE_BEFORE| 0|1410375155145569705| |jack| banana| UPDATE_AFTER| 0|1410375155145569705| |john|pineapple| DELETE| 1|6803657404732109534| +----+---------+-------------+---------------+-------------------+
We can do the same with the minimized CDC view.
+-----+---------+------------+---------------+-------------------+ | name| fruit|_change_type|_change_ordinal|_commit_snapshot_id| +-----+---------+------------+---------------+-------------------+ | jack| apple| INSERT| 0| 260123709935954149| | john|pineapple| INSERT| 0| 260123709935954149| |sarah| orange| INSERT| 0| 260123709935954149| +-----+---------+------------+---------------+-------------------+ +----+---------+------------+---------------+-------------------+ |name| fruit|_change_type|_change_ordinal|_commit_snapshot_id| +----+---------+------------+---------------+-------------------+ |jack| apple| DELETE| 0|1410375155145569705| |jack| banana| INSERT| 0|1410375155145569705| |john|pineapple| DELETE| 1|6803657404732109534| +----+---------+------------+---------------+-------------------+
This time the results in the John row appear in each incremental query. Notice that the Jack row appears twice in the second query, as an INSERT and a DELETE. This is because the compute_updates option that produces the UPDATE_BEFORE and UPDATE_AFTER is not available with net_changes (a minimized CDC view).
That gives you an idea of what is possible with Spark. The major downside here is that because Iceberg does not support materializing CDC files or row lineage, it’s down to the compute engine to infer CDC by reading and comparing snapshots, which is more expensive both in IO and compute. Also note that Iceberg’s incremental changelog scan only supports COW, another major limitation. Once that is fixed, perhaps Spark could use a MOR-compatible CDC inference implementation.
Row-lineage (coming to Iceberg)
It would be nice for Iceberg to offer an append-only change query that returns all inserts, not just inserts from Append snapshots. Row lineage would definitely help implement a fully conformant, efficient append-only change query.
Row lineage or row tracking requires that each row be written with some additional metadata, including:
A row ID that identifies the same row across snapshots.
A row versioning column that indicates to a compute engine when/where the row was first or last inserted. This column will be null for an inserted row.
An append-only change query simply needs to scan added data files for rows that have a null row tracking version column, which is extremely efficient. CDC change queries would use the versioning column to read the delete and update-before rows in their current locations.
There is an early stage, bare-boned design document for row lineage in Apache Iceberg written by Snowflake engineers. Hopefully, row lineage can make it into Iceberg v3.
Conclusions
Current support for Iceberg change queries in Flink, Spark and Spark Structured Streaming:
append-only: Yes, but only Append snapshots, so some inserts can be missed.
Including inserts from MERGE statements would be expensive as it would require additional file scanning and row-level comparisons. Row lineage will solve this problem.
upsert: No support.
There’s nothing stopping compute engines from implementing this, especially for MOR tables.
min-delta: Spark only, via the infer-CDC-on-read approach.
full-delta: Spark only, via the infer-CDC-on-read approach.
The underlying design of the table format will make certain features easier or more difficult for the compute engines to implement efficiently. In the case of append-only change queries, the lack of row lineage makes it inefficient to include the inserts from Overwrite snapshots created by MERGE statements. Hence, no compute engine has implemented it. Likewise, without CDC support, it is more work and less efficient for a compute engine to implement, with only Apache Spark having done so. So Iceberg has some work to do to improve both latest-state (append-only/upsert) and CDC change queries. Row lineage, that may ship in Iceberg v3, will help a lot here.
The question for me is how a row-lineage-based CDC implementation would perform against a “materialize CDC files on write” approach that some other table formats have chosen. The main arguments in favor of row lineage are greater space efficiency and lower writer overhead compared to materializing CDC files. However, given the low cost of S3 storage, I suspect that being more IO and compute efficient for CDC readers is more important than space efficiency, so from my armchair I lean towards CDC file materialization. Snowflake engineers seem pretty certain on row lineage as the way to go.
Things are moving fast, and it would be great if a year from now I could update this post with details on better change query support in Iceberg, especially CDC.